Taxonomic data models
Classifications of organisms and communities provide a complex set of problems that must be addressed in any information system containing references to biological taxa or ecological communities. The core problem is that taxonomic standards vary with time, place, and investigator such that biological taxa and communities frequently have multiple names and those same names frequently have been applied to multiple taxon concepts. When we combine diverse data into a single database, we need to reconcile those different standards.
The traditional solution has been to agree on a standard list and to map all the various applications onto that list. For example, within the US we have several standard lists of plant taxa including Kartesz (1999), USDA PLANTS, ITIS, and NatureServe Explorer. However, these lists fail to allow effective dataset integration for several reasons. (1) Online lists are periodically updated but usually are not simultaneously archived, with the consequence that the user cannot reconstruct the database for some arbitrary time in the past. (2) Ambiguity arises from the fact that one name can be used for multiple taxonomic concepts and one concept can be labeled with multiple names. The standard lists do not define the taxonomic concepts employed, or how they have changed as the list has evolved. (3) Different parties have different perspectives on acceptable names and the meanings associated with them
Part of the ambiguity arises from the requirement of biological nomenclature that when a taxon is split, the name continues to be applied to the taxon that includes the type specimen for the original name. Consider the case of shagbark hickory, which some authors think of as a single entity and others think should be divided into two: northern and southern shagbark. If you encounter the name "Carya ovata (Miller) K. Koch" in a database, you cannot be sure whether it means all shagbarks (sec. Gleason 1952), or just northern shagbark (sec. Radford et al. 1968). Trees that Radford recognizes as Carya carolinae-septentrionalis would be lumped within Carya ovata by a worker who follows Gleason. In addition, authors differ as to whether they believe the two types of shagbark should be recognized as distinct species or simply as varieties. Even if you know a worker follows Strong's treatment of Carya in Flora North America (1997), you cannot be sure whether plants identified as Carya ovata include just Carya ovata var. ovata, (= C. ovata sec. Radford et al.) or also includes C. o. var australis (= Carya carolinae-septentrionalis). The adjacent figure shows the three names (technically four with C.o. var o.) associated with the shagbarks, the three references used above, and how names and references combine to form three concepts. Note that there are two alternative and synonymous name-reference pairs for each of the three concepts.
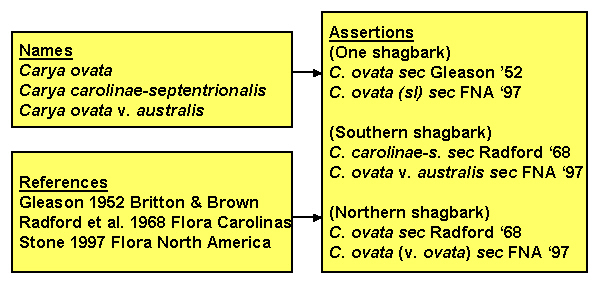
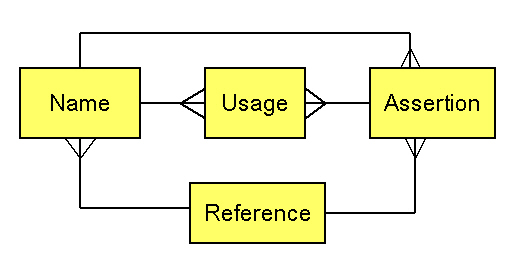
We follow Richard Pyle in referring to the name-reference couplet as an "assertion" (similar in meaning to "potential taxon" as used by Berendsohn 1995). Several points emerge from this example. (1) A name-reference combination constitutes an assertion of a taxonomic concept, though that assertion may be synonymous with other assertions. (2) Identifications (e.g., plot occurrences, specimen labels, treatments in authoritative works) should be by reference to an assertion so as to allow identification of the taxonomic concept intended. (3) A party might choose to recognize a certain set of assertions as standard, and simultaneously recognize other assertions as nonstandard. (4) Identification of the appropriate assertion to attach to an organism does not immediately dictate which name should be applied to that assertion.
We designed a concept-based taxonomy data model and organized with ITIS and the FGDC Biological Data Working Group a meeting to evaluate it. Our model centers on the Assertion as the primary entity for labeling a community or an organism. This is represented in the adjacent figure as an intersection of a Name and a Publication wherein that name was used. An Interpretation occurs when an organism or plot observation is labeled with an assertion. As there can be many assertions that are synonyms or near synonyms, one assertion needs to be chosen as having a Status of standard. All nonstandard assertions should have specifiable Correlations with one or more standard assertions. Similarly, an assertion often has a Lineage such that predecessor and successor standard assertions are tracked. A specific Party (e.g., ITIS, NatureServe, Flora North America) can have its own views as to which assertions as standard, and thus will have its own interpretations as to correlations. Although a name is a critical component of the definition of an assertion, this is not necessarily the name that a party would choose to apply to the assertion. A name Usage is a party-specific application of a name to an assertion, which allows names to change without necessarily changing the assertions tracked in the database.
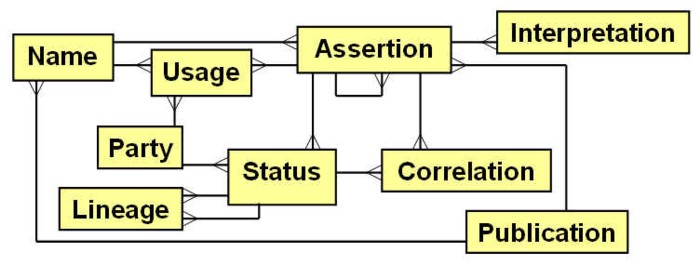
The model we developed is similar to the data model proposed by the International Organization for Plant Information (IOPI; Berendsohn 1997). However, our model incorporates a number of advances over the IOPI model. The core entity in the IOPI model is the "Potential taxon", which represents a combination of our Assertion and Usage entities. Keeping these separate clarifies the important difference between taxonomic concepts and the names applied to those concepts, thereby creating greater stability in the contents of derivative and linked databases. The IOPI model does not explicitly recognize party perspectives, but instead maps preferred views as deriving from publications. Because the IOPI model is organized around fixed publication events, it does not efficiently map continually changing party perspectives, making it difficult to provide software generated time-specific party views of the database.

